AMINO ACID METABOLISM |
• Amino
acids (AAs) are precursors for proteins.
• Precursors
for many other biological N-containing compounds.
• Energy
metabolites: When degraded, amino acids produce glucose, carbohydrates and ketone bodies.
• Excess
dietary AAs are neither stored nor excreted. Rather, they are converted to common
metabolic intermediates.
Fate of Amino Group
1. Ureotelic: urea
for excretion most
terrestrial vertebrates
2. Uricotelic: uric
acid for excretion birds,
reptiles
3. Ammonotelic: NH4+
for excretion aquatic
animals
Fate of Carbon Skeletons
Converted into 7 common metabolites:
• pyruvate; • acetyl-CoA; •
acetoacetate; •
a-ketoglutarate;
• succinyl-CoA; • fumarate; • oxaloacetate
|
FATE OF AMINO GROUP |
I. DEAMINATION
A. Transamination by Aminotransferase (or Transaminase)
• Funnel
a-amino
groups from a variety of AAs to glutamate by reacting
with a-ketoglutarate.
amino acid + a-ketoglutarate
⇌ a-keto
acid + glutamate
· Does not result in any net deamination.
B. Oxidative Deamination
1. Glutamate Dehydrogenase (in
mitochondria)
·
See
p.692
·
Glu
+ NAD+ (or NADP+) + H2O ⇌
NH4+ + a-ketoglutarate
+ NAD(P)H +H+
·
An enzyme unusual (but not the only one as stated in the Textbook)
in being able to use NAD+ and NADP+.
·
Plays
a central role in AA metabolism. In most
organisms glutamate is the only AA which has such an oxidative deamination enzyme.
·
Glutamate
DH is allosterically regulated. It is inhibited
by GTP and ATP, and activated by GDP and ADP.
·
The
NH4+ so obtained can feed into
urea cycle.
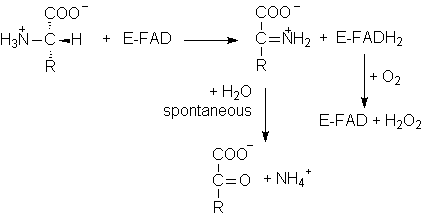
2. L-Amino Acid Oxidase
·
Requires
FAD as a cofactor.
·
D-Amino
acid oxidase also exists in mammalian tissues.
Real physiological function unknown.
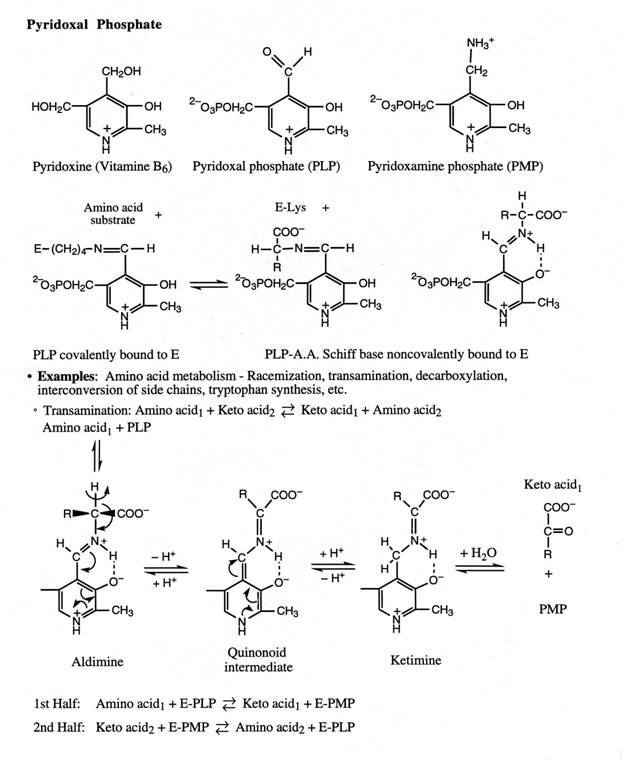
C. Direct Deamination of Serine
and Histidine
1. Serine Dehydratase
·
Fig.
20-15.
·
PLP-dependent
·
serine
+ H2O ® pyruvate
+ NH4+
2. Histidine Ammonia Lyase
·
Fig.
20-17, Reaction 8.
·
histidine
® urocanate + NH4+
|
UREA
CYCLE |
·
1932
by Hans Krebs and Kurt Henseleit as the first
metabolic cycle elucidated. See Fig.
20-9.
·
Overall
Reaction:
·
NH3 + HCO3– +
aspartate + 3 ATP + H2O ® urea + fumarate + 2 ADP + 2 Pi +
AMP + PPi
·
Requires
5 enzymes: 2 from mitochondria and 3
from cytosol.
1. Carbamoyl phosphate synthetase
(Mitochondrial)
·
Eukaryotes
have two forms of CPS, the mitochondrial CPS I uses ammonia as the N donor for
urea synthesis. The cytosolic CPS II
uses glutamine as its N donor for pyrimidine
biosynthesis.
·
2
ATP + HCO3– + NH3 ® carbamoyl
phosphate + 2 ADP + Pi
2. Ornithine transcarbamoylase
(Mitochondrial)
·
carbamoyl phosphate + ornithine ® citrulline
Antiport: (cytosolic ornithine ® mitochondria) coupled to (mitochondrial citrulline
® cytosol).
3. Argininosuccinate synthetase
(Cytosolic)
·
citrulline + aspartate + ATP ® argininosuccinate
+ AMP + PPi
4. Argininosuccinase (Cytosolic)
·
argininosuccinate ® fumarate
+ arginine
·
The
skeleton of Asp is recovered in fumarate.
Up to this point, the reactions are the same for all organisms that are
capable of synthesizing arginine.
5. Arginase (Cytosolic)
·
Only
the ureotelic animals have large amounts of the arginase.
·
arginine
+ H2O ® urea + ornithine
|
REGULATION OF UREA
CYCLE |
1. Mitochondrial carbamoyl
phosphate synthetase I (CPS I)
·
CPS
I catalyzes the first committed step of the urea cycle.
·
CPS
I is also an allosteric enzyme sensitive to activation
by N-acetylglutamate which is derived from glutamate
and acetyl-CoA.
·
Increased
rate of AA degradation requires higher rate of urea synthesis.
·
AA degradation ® ↑glutamate
concentration → ↑synthesis
of N-acetylglutamate ® ↑CPS
I activity ® ↑urea cycle efficiency
2. All other urea cycle enzymes are controlled by the concentrations
of their substrates.
·
Deficiency
in an E ® ↑(substrate) ® ↑rate of the deficient E.